Research Projects (FY2023-2024)
Understanding chromosome condensation during mitotic phase through the mode coupling between bend and twist
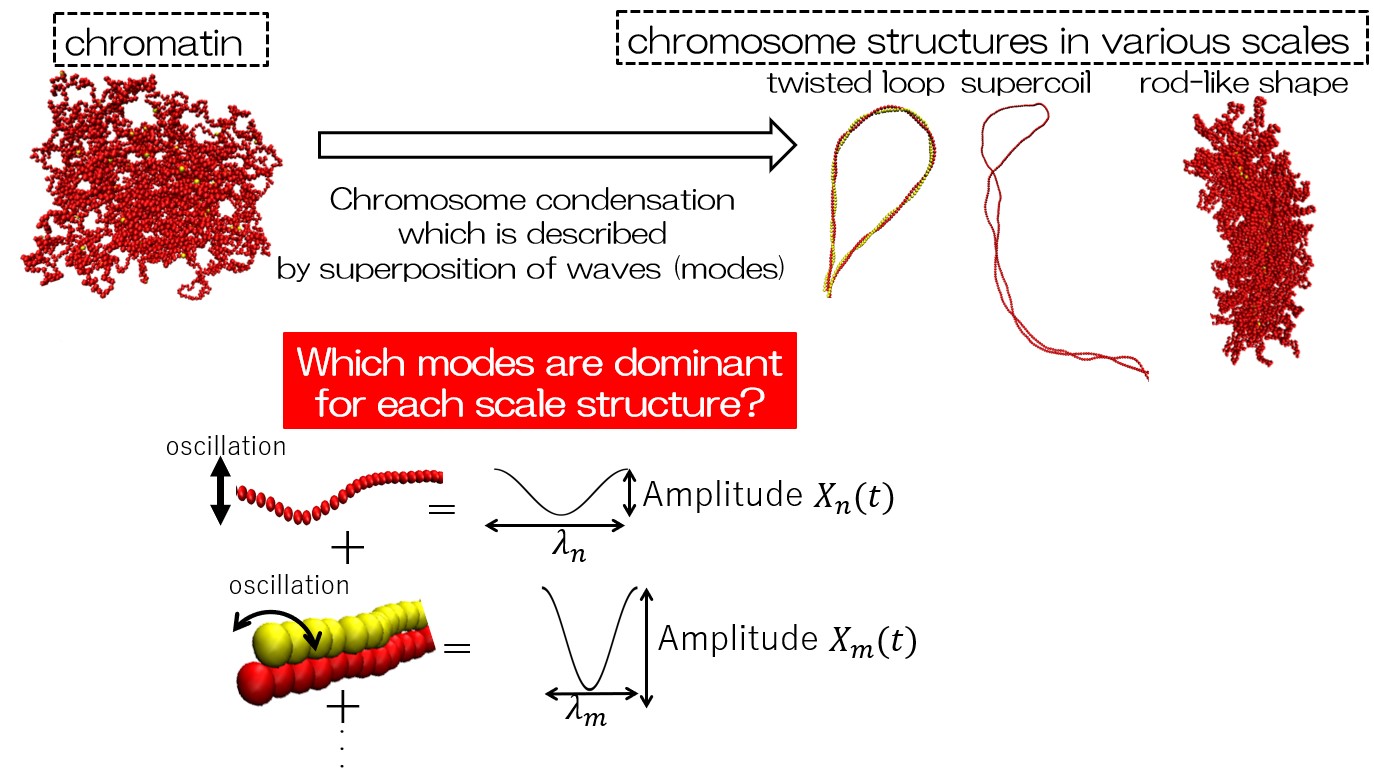
In twistable chains such as DNA or chromatin, the loop formation dynamics is expected to show various motions in different temporal-spatial scales, and thereby to construct different structures over the different scales. Polymer dynamics can be described by the superposition of dynamical modes (waves running along the chain) or the coupling of these modes. The relaxation time of the amplitude of the mode specifies the time scale of the component of the dynamics, while the wavelength indicates the spatial scale. In this research, we theoretically clarify which modes play an important role on the loop formation dynamics in chromosome condensation by decomposing model chain dynamics in the mechanical model into each scale component.

Hiroshi Yokota Project Leader
- Graduate School of Human and Environmental Studies, Kyoto University

